DNA helicases Sgs1 and BLM promote DNA double-strand break resection
- PMID: 18923075
- PMCID: PMC2569880
- DOI: 10.1101/gad.503108
DNA helicases Sgs1 and BLM promote DNA double-strand break resection
Abstract
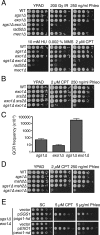
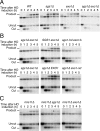
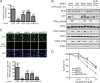
A key cellular response to DNA double-strand breaks (DSBs) is 5'-to-3' DSB resection by nucleases to generate regions of ssDNA that then trigger cell cycle checkpoint signaling and DSB repair by homologous recombination (HR). Here, we reveal that in the absence of exonuclease Exo1 activity, deletion or mutation of the Saccharomyces cerevisiae RecQ-family helicase, Sgs1, causes pronounced hypersensitivity to DSB-inducing agents. Moreover, we establish that this reflects severely compromised DSB resection, deficient DNA damage signaling, and strongly impaired HR-mediated repair. Furthermore, we show that the mammalian Sgs1 ortholog, BLM--whose deficiency causes cancer predisposition and infertility in people--also functions in parallel with Exo1 to promote DSB resection, DSB signaling and resistance to DSB-generating agents. Collectively, these data establish evolutionarily conserved roles for the BLM and Sgs1 helicases in DSB processing, signaling, and repair.
Figures
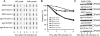
Comment in
-
DNA double-strand break processing: the beginning of the end.Genes Dev. 2008 Nov 1;22(21):2903-7. doi: 10.1101/gad.1742408. Genes Dev. 2008. PMID: 18981468 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
