Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs
- PMID: 18923076
- PMCID: PMC2569885
- DOI: 10.1101/gad.1705308
Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs
Abstract
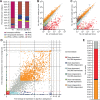
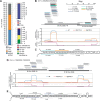
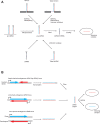
Canonical microRNAs (miRNAs) require two processing steps: the first by the Microprocessor, a complex of DGCR8 and Drosha, and the second by a complex of TRBP and Dicer. dgcr8Delta/Delta mouse embryonic stem cells (mESCs) have less severe phenotypes than dicer1Delta/Delta mESCs, suggesting a physiological role for Microprocessor-independent, Dicer-dependent small RNAs. To identify these small RNAs with unusual biogenesis, we performed high-throughput sequencing from wild-type, dgcr8Delta/Delta, and dicer1Delta/Delta mESCs. Several of the resulting DGCR8-independent, Dicer-dependent RNAs were noncanonical miRNAs. These derived from mirtrons and a newly identified subclass of miRNA precursors, which appears to be the endogenous counterpart of shRNAs. Our analyses also revealed endogenous siRNAs resulting from Dicer cleavage of long hairpins, the vast majority of which originated from one genomic locus with tandem, inverted short interspersed nuclear elements (SINEs). Our results extend the known diversity of mammalian small RNA-generating pathways and show that mammalian siRNAs exist in cell types other than oocytes.
Figures


References
-
- Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. - PubMed
-
- Altman S. The road to RNase P. Nat. Struct. Biol. 2000;7:827–828. - PubMed
-
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. - PubMed
-
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
