Ion channels in microbes
- PMID: 18923187
- PMCID: PMC2579964
- DOI: 10.1152/physrev.00005.2008
Ion channels in microbes
Abstract
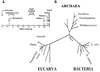
Studies of ion channels have for long been dominated by the animalcentric, if not anthropocentric, view of physiology. The structures and activities of ion channels had, however, evolved long before the appearance of complex multicellular organisms on earth. The diversity of ion channels existing in cellular membranes of prokaryotes is a good example. Although at first it may appear as a paradox that most of what we know about the structure of eukaryotic ion channels is based on the structure of bacterial channels, this should not be surprising given the evolutionary relatedness of all living organisms and suitability of microbial cells for structural studies of biological macromolecules in a laboratory environment. Genome sequences of the human as well as various microbial, plant, and animal organisms unambiguously established the evolutionary links, whereas crystallographic studies of the structures of major types of ion channels published over the last decade clearly demonstrated the advantage of using microbes as experimental organisms. The purpose of this review is not only to provide an account of acquired knowledge on microbial ion channels but also to show that the study of microbes and their ion channels may also hold a key to solving unresolved molecular mysteries in the future.
Figures

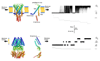



References
-
- Ajouz B, Berrier C, Besnard M, Martinac B, Ghazi A. Contributions of the different extramembranous domains of the mechanosensitive ion channel MscL to its response to membrane tension. J Biol Chem. 2000;275:1015–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

