Dynamic protein methylation in chromatin biology
- PMID: 18923809
- PMCID: PMC2794343
- DOI: 10.1007/s00018-008-8303-z
Dynamic protein methylation in chromatin biology
Abstract
Post-translational modification of chromatin is emerging as an increasingly important regulator of chromosomal processes. In particular, histone lysine and arginine methylation play important roles in regulating transcription, maintaining genomic integrity, and contributing to epigenetic memory. Recently, the use of new approaches to analyse histone methylation, the generation of genetic model systems, and the ability to interrogate genome wide histone modification profiles has aided in defining how histone methylation contributes to these processes. Here we focus on the recent advances in our understanding of the histone methylation system and examine how dynamic histone methylation contributes to normal cellular function in mammals.
Figures

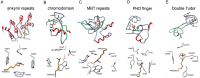

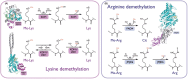

References
-
- Kossel A. Ueber einen peptoartigen bestandheil des zellkerns. Z. Physiol. Chem. 1884;8:511–515.
-
- Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
-
- Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. - PubMed
-
- Chakravarthy S., Park Y.J., Chodaparambil J., Edayathumangalam R.S., Luger K. Structure and dynamic properties of nucleosome core particles. FEBS Lett. 2005;579:895–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

