Parallel analysis of tetramerization domain mutants of the human p53 protein using PCR colonies
- PMID: 18923936
- PMCID: PMC2269032
- DOI: 10.1007/s11568-007-9011-8
Parallel analysis of tetramerization domain mutants of the human p53 protein using PCR colonies
Abstract
A highly-parallel yeast functional assay, capable of screening approximately 100-1,000 mutants in parallel and designed to screen the activity of transcription activator proteins, was utilized to functionally characterize tetramerization domain mutants of the human p53 transcription factor and tumor suppressor protein. A library containing each of the 19 possible single amino acid substitutions (57 mutants) at three positions in the tetramerization domain of the human p53 protein, was functionally screened in Saccharomyces cerevisiae. Amino acids Leu330 and Ile332, whose side chains form a portion of a hydrophobic pocket that stabilizes the active p53 tetramer, were found to tolerate most hydrophobic amino acid substitutions while hydrophilic substitutions resulted in the inactivation of the protein. Amino acid Gln331 tolerated essentially all mutations. Importantly, highly parallel mutagenesis and cloning techniques were utilized which, in conjunction with recently reported highly parallel DNA sequencing methods, would be capable of increasing throughput an additional 2-3 orders of magnitude.
Figures

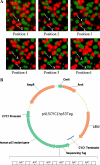

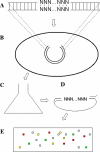
References
-
- Bitter GA, Schaeffer TN, Ellison AR (2002) Reporter gene regulation in Saccharomyces cerevisiae by the human p53 tumor suppressor protein. J Mol Microbiol Biotechnol 4(6):539–550 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
