Tau isoform regulation is region- and cell-specific in mouse brain
- PMID: 18925637
- PMCID: PMC2845852
- DOI: 10.1002/cne.21867
Tau isoform regulation is region- and cell-specific in mouse brain
Abstract
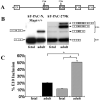
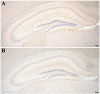
Tau is a microtubule-associated protein implicated in neurodegenerative tauopathies. Alternative splicing of the tau gene (MAPT) generates six tau isoforms, distinguishable by the exclusion or inclusion of a repeat region of exon 10, which are referred to as 3-repeat (3R) and 4-repeat (4R) tau, respectively. We developed transgenic mouse models that express the entire human MAPT gene in the presence and absence of the mouse Mapt gene and compared the expression and regulation of mouse and human tau isoforms during development and in the young adult. We found differences between mouse and human tau in the regulation of exon 10 inclusion. Despite these differences, the isoform splicing pattern seen in normal human brain is replicated in our mouse models. In addition, we found that all tau, both in the neonate and young adult, is phosphorylated. We also examined the normal anatomic distribution of mouse and human tau isoforms in mouse brain. We observed developmental and species-specific variations in the expression of 3R- and 4R-tau within the frontal cortex and hippocampus. In addition, there were differences in the cellular distribution of the isoforms. Mice transgenic for the human MAPT gene exhibited higher levels of neuronal cell body expression of tau compared to wildtype mice. This neuronal cell body expression of tau was limited to the 3R isoform, whereas expression of 4R-tau was more "synaptic like," with granular staining of neuropil rather than in neuronal cell bodies. These developmental and species-specific differences in the regulation and distribution of tau isoforms may be important to the understanding of normal and pathologic tau isoform expression.
Figures
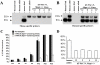




References
-
- Andorfer C, Kress Y, Espinoza M, de Silva R, Tucker KL, Barde YA, Duff K, Davies P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 2003;86:582–590. - PubMed
-
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim. Biophys. Acta. 2005;1739:91–103. - PubMed
-
- Arai T, Ikeda K, Akiyama H, Shikamoto Y, Tsuchiya K, Yagishita S, Beach T, Rogers J, Schwab C, Mcgeer PL. Distinct isoforms of tau aggregated in neurons and glial cells in brains of patients with Pick’s disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropath. 2001;101:167–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

