Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes
- PMID: 18925961
- PMCID: PMC2588460
- DOI: 10.1186/1471-2407-8-299
Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes
Abstract
Background: Chronic myelomonocytic leukemia (CMML) is a hematological disease close to, but separate from both myeloproliferative disorders (MPD) and myelodysplastic syndromes and may show either myeloproliferative (MP-CMML) or myelodysplastic (MD-CMML) features. Not much is known about the molecular biology of this disease.
Methods: We studied a series of 30 CMML samples (13 MP- and 11 MD-CMMLs, and 6 acutely transformed cases) from 29 patients by using Agilent high density array-comparative genomic hybridization (aCGH) and sequencing of 12 candidate genes.
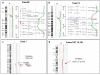
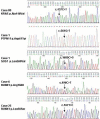
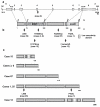
Results: Two-thirds of samples did not show any obvious alteration of aCGH profiles. In one-third we observed chromosome abnormalities (e.g. trisomy 8, del20q) and gain or loss of genes (e.g. NF1, RB1 and CDK6). RAS mutations were detected in 4 cases (including an uncommon codon 146 mutation in KRAS) and PTPN11 mutations in 3 cases. We detected 11 RUNX1 alterations (9 mutations and 2 rearrangements). The rearrangements were a new, cryptic inversion of chromosomal region 21q21-22 leading to break and fusion of RUNX1 to USP16. RAS and RUNX1 alterations were not mutually exclusive. RAS pathway mutations occurred in MP-CMMLs (approximately 46%) but not in MD-CMMLs. RUNX1 alterations (mutations and cryptic rearrangement) occurred in both MP and MD classes (~38%).
Conclusion: We detected RAS pathway mutations and RUNX1 alterations. The latter included a new cryptic USP16-RUNX1 fusion. In some samples, two alterations coexisted already at this early chronic stage.
Figures


References
-
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick H, Sultan C, Cox C. The chronic myeloid leukaemias: guidelines for distinguishing chronic granulocytic, atypical chronic myeloid, and chronic myelomonocytic leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br J Haematol. 1994;84:746–754. doi: 10.1111/j.1365-2141.1994.tb06734.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

