Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance
- PMID: 18926708
- PMCID: PMC2606042
- DOI: 10.1016/j.tibs.2008.09.003
Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance
Abstract
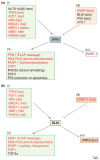
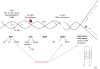
The RecQ helicases are guardians of the genome. Members of this conserved family of proteins have a key role in protecting and stabilizing the genome against deleterious changes. Deficiencies in RecQ helicases can lead to high levels of genomic instability and, in humans, to premature aging and increased susceptibility to cancer. Their diverse roles in DNA metabolism, which include a role in telomere maintenance, reflect interactions with multiple cellular proteins, some of which are multifunctional and also have very diverse functions. The results of in vitro cellular and biochemical studies have been complimented by recent in vivo studies using genetically modified mouse strains. Together, these approaches are helping to unravel the mechanism(s) of action and biological functions of the RecQ helicases.
Figures





References
-
- German J. Bloom’s syndrome. XX The first 100 cancers. Cancer Genet Cytogenet. 1997;93:100–106. - PubMed
-
- Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. - PubMed
-
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. - PubMed
-
- von Kobbe C, et al. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–22044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

