Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1
- PMID: 18926831
- PMCID: PMC2631168
- DOI: 10.1016/j.jmb.2008.09.065
Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1
Abstract
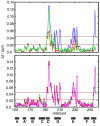
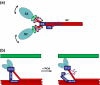
Myosin binding protein C (MyBP-C) is a thick filament protein involved in the regulation of muscle contraction. Mutations in the gene for MyBP-C are the second most frequent cause of hypertrophic cardiomyopathy. MyBP-C binds to myosin with two binding sites, one at its C-terminus and another at its N-terminus. The N-terminal binding site, consisting of immunoglobulin domains C1 and C2 connected by a flexible linker, interacts with the S2 segment of myosin in a phosphorylation-regulated manner. It is assumed that the function of MyBP-C is to act as a tether that fixes the S1 heads in a resting position and that phosphorylation releases the S1 heads into an active state. Here, we report the structure and binding properties of domain C1. Using a combination of site-directed mutagenesis and NMR interaction experiments, we identified the binding site of domain C1 in the immediate vicinity of the S1-S2 hinge, very close to the light chains. In addition, we identified a zinc binding site on domain C1 in close proximity to the S2 binding site. Its zinc binding affinity (K(d) of approximately 10-20 microM) might not be sufficient for a physiological effect. However, the familial hypertrophic cardiomyopathy-related mutation of one of the zinc ligands, glutamine 210 to histidine, will significantly increase the binding affinity, suggesting that this mutation may affect S2 binding. The close proximity of the C1 binding site to the hinge, the light chains and the S1 heads also provides an explanation for recent observations that (a) shorter fragments of MyBP-C unable to act as a tether still have an effect on the actomyosin ATPase and (b) as to why the myosin head positions in phosphorylated wild-type mice and MyBP-C knockout mice are so different: Domain C1 bound to the S1-S2 hinge is able to manipulate S1 head positions, thus influencing force generation without tether. The potentially extensive extra interactions of C1 are expected to keep it in place, while phosphorylation dislodges the C1-C2 linker and domain C2. As a result, the myosin heads would always be attached to a tether that has phosphorylation-dependent length regulation.
Figures







References
-
- Kobayashi T., Solaro R.J. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005;67:39–67. - PubMed
-
- Winegrad S. Myosin-binding protein C (MyBP-C) in cardiac muscle and contractility. Adv. Exp. Med. Biol. 2003;538:31–40. Discussion 40-1. - PubMed
-
- Oakley C.E., Hambly B.D., Curmi P.M.G., Brown L.J. Myosin binding protein C: structural abnormalities in familial hypertrophic cardiomyopathy. Cell Res. 2004;14:95–110. - PubMed
-
- Winegrad S. Myosin binding protein C, a potential regulator of cardiac contractility. Circulation. 2000;86:6–7. - PubMed
-
- Lange S., Ehler E., Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

