Characterization of reticulate networks based on the coalescent with recombination
- PMID: 18927089
- PMCID: PMC2582979
- DOI: 10.1093/molbev/msn219
Characterization of reticulate networks based on the coalescent with recombination
Abstract
Phylogenetic networks aim to represent the evolutionary history of taxa. Within these, reticulate networks are explicitly able to accommodate evolutionary events like recombination, hybridization, or lateral gene transfer. Although several metrics exist to compare phylogenetic networks, they make several assumptions regarding the nature of the networks that are not likely to be fulfilled by the evolutionary process. In order to characterize the potential disagreement between the algorithms and the biology, we have used the coalescent with recombination to build the type of networks produced by reticulate evolution and classified them as regular, tree sibling, tree child, or galled trees. We show that, as expected, the complexity of these reticulate networks is a function of the population recombination rate. At small recombination rates, most of the networks produced are already more complex than regular or tree sibling networks, whereas with moderate and large recombination rates, no network fit into any of the standard classes. We conclude that new metrics still need to be devised in order to properly compare two phylogenetic networks that have arisen from reticulating evolutionary process.
Figures
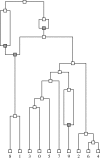
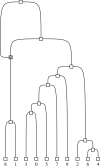
References
-
- Baroni M, Semple C, Steel M. A framework for representing reticulate evolution. Ann Combin. 2004;8:391–408.
-
- Cardona G, Llabrés M, Rosselló F, Valiente G Forthcoming. Metrics for phylogenetic networks I: generalizations of the Robinson-Foulds metric. IEEE/ACM Trans Comput Biol Bioinform. 2008b - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

