A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo
- PMID: 18927239
- PMCID: PMC5419408
- DOI: 10.1210/me.2008-0097
A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo
Abstract
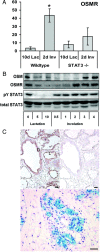
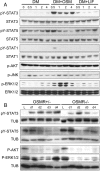
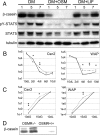
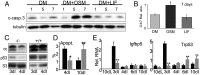
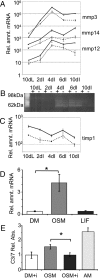
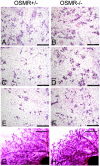
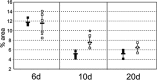
Recent studies in breast cancer cell lines have shown that oncostatin M (OSM) not only inhibits proliferation but also promotes cell detachment and enhances cell motility. In this study, we have looked at the role of OSM signaling in nontransformed mouse mammary epithelial cells in vitro using the KIM-2 mammary epithelial cell line and in vivo using OSM receptor (OSMR)-deficient mice. OSM and its receptor were up-regulated approximately 2 d after the onset of postlactational mammary regression, in response to leukemia inhibitory factor (LIF)-induced signal transducer and activator of transcription-3 (STAT3). This resulted in sustained STAT3 activity, increased epithelial apoptosis, and enhanced clearance of epithelial structures during the remodeling phase of mammary involution. Concurrently, OSM signaling precipitated the dephosphorylation of STAT5 and repressed expression of the milk protein genes beta-casein and whey acidic protein (WAP). Similarly, during pregnancy, OSM signaling suppressed beta-casein and WAP gene expression. In vitro, OSM but not LIF persistently down-regulated phosphorylated (p)-STAT5, even in the continued presence of prolactin. OSM also promoted the expression of metalloproteinases MMP3, MMP12, and MMP14, which, in vitro, were responsible for OSM-specific apoptosis. Thus, the sequential activation of IL-6-related cytokines during mammary involution culminates in an OSM-dependent repression of epithelial-specific gene expression and the potentiation of epithelial cell extinction mediated, at least in part, by the reciprocal regulation of p-STAT5 and p-STAT3.
Figures
References
-
- Gearing DP 1993. The leukemia inhibitory factor and its receptor. Adv Immunol 53:31–58 - PubMed
-
- Tanaka M, Miyajima A 2003. Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol 149:39–52 - PubMed
-
- Bruce AG, Hoggatt IH, Rose TM 1992. Oncostatin M is a differentiation factor for myeloid leukemia cells. J Immunol 149:1271–1275 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

