Surface sites for engineering allosteric control in proteins
- PMID: 18927392
- PMCID: PMC3071530
- DOI: 10.1126/science.1159052
Surface sites for engineering allosteric control in proteins
Abstract
Statistical analyses of protein families reveal networks of coevolving amino acids that functionally link distantly positioned functional surfaces. Such linkages suggest a concept for engineering allosteric control into proteins: The intramolecular networks of two proteins could be joined across their surface sites such that the activity of one protein might control the activity of the other. We tested this idea by creating PAS-DHFR, a designed chimeric protein that connects a light-sensing signaling domain from a plant member of the Per/Arnt/Sim (PAS) family of proteins with Escherichia coli dihydrofolate reductase (DHFR). With no optimization, PAS-DHFR exhibited light-dependent catalytic activity that depended on the site of connection and on known signaling mechanisms in both proteins. PAS-DHFR serves as a proof of concept for engineering regulatory activities into proteins through interface design at conserved allosteric sites.
Figures



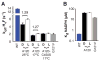
References
-
- Richards FM, Lim WA. Q Rev Biophys. 1993;26:423. - PubMed
-
- Tsai J, Taylor R, Chothia C, Gerstein M. J Mol Biol. 1999;290:253. - PubMed
-
- Swain JF, Gierasch LM. Curr Opin Struct Biol. 2006;16:102. - PubMed
-
- Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196. - PubMed
-
- Hammes-Schiffer S, Benkovic SJ. Annu Rev Biochem. 2006;75:519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

