Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry
- PMID: 18927556
- PMCID: PMC2860270
- DOI: 10.1038/nprot.2008.159
Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry
Abstract
Advancement in proteomics research relies on the development of new, innovative tools for identifying and characterizing proteins. Here, we describe a protocol for analyzing peptides and proteins on a chromatographic timescale by coupling nanoflow reverse-phase (RP) liquid chromatography (LC) to electron-transfer dissociation (ETD) mass spectrometry. For this protocol, proteins can be proteolytically digested before ETD analysis, although digestion is not necessary for all applications. Proteins <or=30 kDa can be analyzed intact, particularly if the objective is protein identification. Peptides or proteins are loaded onto a RP column and are gradient-eluted into an ETD-enabled mass spectrometer. ETD tandem mass spectrometry (MS/MS) provides extensive sequence information required for the unambiguous identification of peptides and proteins and for characterization of posttranslational modifications. ETD is a powerful MS/MS technique and does not compromise the sensitivity and speed necessary for online chromatographic separations. The described procedure for sample preparation, column packing, sample loading and ETD analysis can be implemented in 5-15 h.
Figures


 represents a phosphate group and is positioned above the phosphorylated serine residue. A triangle (▼) is positioned above m/z peaks that are within the precursor isolation window. Charge-reduced species and species resulting from neutral losses are bracketed.
represents a phosphate group and is positioned above the phosphorylated serine residue. A triangle (▼) is positioned above m/z peaks that are within the precursor isolation window. Charge-reduced species and species resulting from neutral losses are bracketed.
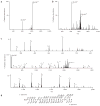

 is positioned above the arginine residue found to be dimethylated. A triangle (▼) is positioned above m/z peaks that are within the precursor isolation window.
is positioned above the arginine residue found to be dimethylated. A triangle (▼) is positioned above m/z peaks that are within the precursor isolation window.
References
-
- Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. - PubMed
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Yamashita M, Fenn JB. Electrospray ion source. Another variation on the free-jet theme. J Phys Chem. 1984;88:4451–4459.
-
- Eng JK, McCormack AL, Yates I, John R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
-
- Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

