Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail
- PMID: 18927579
- PMCID: PMC2715827
- DOI: 10.1038/nrg2438
Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail
Abstract
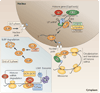
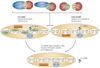
The canonical histone proteins are encoded by replication-dependent genes and must rapidly reach high levels of expression during S phase. In metazoans the genes that encode these proteins produce mRNAs that, instead of being polyadenylated, contain a unique 3' end structure. By contrast, the synthesis of the variant, replication-independent histones, which are encoded by polyadenylated mRNAs, persists outside of S phase. Accurate positioning of both histone types in chromatin is essential for proper transcriptional regulation, the demarcation of heterochromatic boundaries and the epigenetic inheritance of gene expression patterns. Recent results suggest that the coordinated synthesis of replication-dependent and variant histone mRNAs is achieved by signals that affect formation of the 3' end of the replication-dependent histone mRNAs.
Figures


References
-
- Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 2005;21:133–153. - PubMed
-
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. - PubMed
-
- De KL, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nature Struct. Mol. Biol. 2007;14:997–1007. - PubMed
-
- Marzluff WF. Metazoan replication dependent histone mRNAs: a unique class of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

