High-resolution functional profiling of hepatitis C virus genome
- PMID: 18927624
- PMCID: PMC2564836
- DOI: 10.1371/journal.ppat.1000182
High-resolution functional profiling of hepatitis C virus genome
Abstract
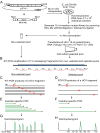
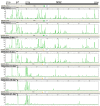
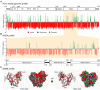
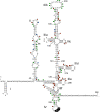
Hepatitis C virus is a leading cause of human liver disease worldwide. Recent discovery of the JFH-1 isolate, capable of infecting cell culture, opens new avenues for studying HCV replication. We describe the development of a high-throughput, quantitative, genome-scale, mutational analysis system to study the HCV cis-elements and protein domains that are essential for virus replication. An HCV library with 15-nucleotide random insertions was passaged in cell culture to examine the effect of insertions at each genome location by insertion-specific fluorescent-PCR profiling. Of 2399 insertions identified in 9517 nucleotides of the genome, 374, 111, and 1914 were tolerated, attenuating, and lethal, respectively, for virus replication. Besides identifying novel functional domains, this approach confirmed other functional domains consistent with previous studies. The results were validated by testing several individual mutant viruses. Furthermore, analysis of the 3' non-translated variable region revealed a spacer role in virus replication, demonstrating the utility of this approach for functional discovery. The high-resolution functional profiling of HCV domains lays the foundation for further mechanistic studies and presents new therapeutic targets as well as topological information for designing vaccine candidates.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. - PubMed
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
-
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5 ed. Philadelphia, PA: Lippincott-Raven Publishers; 2007. pp. 1101–1152.

