Rapid generation of splicing reporters with pSpliceExpress
- PMID: 18930792
- PMCID: PMC2821805
- DOI: 10.1016/j.gene.2008.09.021
Rapid generation of splicing reporters with pSpliceExpress
Abstract
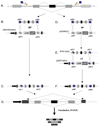
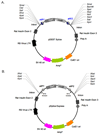
Almost all human protein-coding transcripts undergo pre-mRNA splicing and a majority of them is alternatively spliced. The most common technique used to analyze the regulation of an alternative exon is through reporter minigene constructs. However, their construction is time-consuming and is often complicated by the limited availability of appropriate restriction sites. Here, we report a fast and simple recombination-based method to generate splicing reporter genes, using a new vector, pSpliceExpress. The system allows generation of minigenes within one week. Minigenes generated with pSpliceExpress show the same regulation as displayed by conventionally cloned reporter constructs and provide an alternate avenue to study splice site selection in vivo.
Figures



References
-
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. - PubMed
-
- Cáceres J, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. - PubMed
-
- Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Moller HJ, Andreadis A, Henkel K, Stamm S. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer's disease. J. Neurochem. 2006;96:635–644. - PubMed
-
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

