Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction
- PMID: 18930918
- PMCID: PMC2596406
- DOI: 10.1074/jbc.M806479200
Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction
Abstract
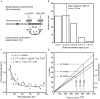
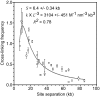
The higher order arrangement of nucleosomes and the level of compaction of the chromatin fiber play important roles in the control of gene expression and other genomic activities. Analysis of chromatin in vitro has suggested that under near physiological conditions chromatin fibers can become highly compact and that the level of compaction can be modulated by histone modifications. However, less is known about the organization of chromatin fibers in living cells. Here, we combine chromosome conformation capture (3C) data with distance measurements and polymer modeling to determine the in vivo mass density of a transcriptionally active 95-kb GC-rich domain on chromosome III of the yeast Saccharomyces cerevisiae. In contrast to previous reports, we find that yeast does not form a compact fiber but that chromatin is extended with a mass per unit length that is consistent with a rather loose arrangement of nucleosomes. Analysis of 3C data from a neighboring AT-rich chromosomal domain indicates that chromatin in this domain is more compact, but that mass density is still well below that of a canonical 30 nm fiber. Our approach should be widely applicable to scale 3C data to real spatial dimensions, which will facilitate the quantification of the effects of chromatin modifications and transcription on chromatin fiber organization.
Figures


References
-
- Belmont, A. S., Dietzel, S., Nye, A. C., Strukov, Y. G., and Tumbar, T. (1999) Curr. Opin. Cell Biol. 11 307–311 - PubMed
-
- Widom, J. (1989) Annu. Rev. Biophys. Biophys. Chem. 18 365–395 - PubMed
-
- van Holde, K. E. (1989) Chromatin, Springer, Heidelberg
-
- van Holde, K., and Zlatanova, J. (1995) J. Biol. Chem. 270 8373–8376 - PubMed
-
- Woodcock, C. L., and Dimitrov, S. (2001) Curr. Opin. Genet. Dev. 11 130–135 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

