The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila
- PMID: 18931655
- PMCID: PMC2597674
- DOI: 10.1038/nature07388
The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila
Abstract
In all animals, the initial events of embryogenesis are controlled by maternal gene products that are deposited into the developing oocyte. At some point after fertilization, control of embryogenesis is transferred to the zygotic genome in a process called the maternal-to-zygotic transition. During this time, many maternal RNAs are degraded and transcription of zygotic RNAs ensues. There is a long-standing question as to which factors regulate these events. The recent findings that microRNAs and Smaug mediate maternal transcript degradation have shed new light on this aspect of the problem. However, the transcription factor(s) that activate the zygotic genome remain elusive. The discovery that many of the early transcribed genes in Drosophila share a cis-regulatory heptamer motif, CAGGTAG and related sequences, collectively referred to as TAGteam sites raised the possibility that a dedicated transcription factor could interact with these sites to activate transcription. Here we report that the zinc-finger protein Zelda (Zld; Zinc-finger early Drosophila activator) binds specifically to these sites and is capable of activating transcription in transient transfection assays. Mutant embryos lacking zld are defective in cellular blastoderm formation, and fail to activate many genes essential for cellularization, sex determination and pattern formation. Global expression profiling confirmed that Zld has an important role in the activation of the early zygotic genome and suggests that Zld may also regulate maternal RNA degradation during the maternal-to-zygotic transition.
Figures

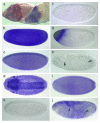
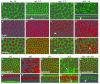
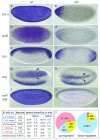
References
-
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. - PubMed
-
- Giraldez AJ, et al. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. - PubMed
-
- Bushati N, Stark A, Brennecke J, Cohen S. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. - PubMed
-
- Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell. 2007;12:143–155. - PubMed
-
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

