IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis
- PMID: 18931663
- PMCID: PMC2818601
- DOI: 10.1038/ncb1789
IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis
Abstract
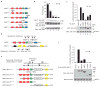
The covalent attachment of ubiquitin to target proteins influences various cellular processes, including DNA repair, NF-kappaB signalling and cell survival. The most common mode of regulation by ubiquitin-conjugation involves specialized ubiquitin-binding proteins that bind to ubiquitylated proteins and link them to downstream biochemical processes. Unravelling how the ubiquitin-message is recognized is essential because aberrant ubiquitin-mediated signalling contributes to tumour formation. Recent evidence indicates that inhibitor of apoptosis (IAP) proteins are frequently overexpressed in cancer and their expression level is implicated in contributing to tumorigenesis, chemoresistance, disease progression and poor patient-survival. Here, we have identified an evolutionarily conserved ubiquitin-associated (UBA) domain in IAPs, which enables them to bind to Lys 63-linked polyubiquitin. We found that the UBA domain is essential for the oncogenic potential of cIAP1, to maintain endothelial cell survival and to protect cells from TNF-alpha-induced apoptosis. Moreover, the UBA domain is required for XIAP and cIAP2-MALT1 to activate NF-kappaB. Our data suggest that the UBA domain of cIAP2-MALT1 stimulates NF-kappaB signalling by binding to polyubiquitylated NEMO. Significantly, 98% of all cIAP2-MALT1 fusion proteins retain the UBA domain, suggesting that ubiquitin-binding contributes to the oncogenic potential of cIAP2-MALT1 in MALT lymphoma. Our data identify IAPs as ubiquitin-binding proteins that contribute to ubiquitin-mediated cell survival, NF-kappaB signalling and oncogenesis.
Figures


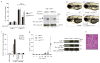

References
-
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nature Rev Mol Cell Biol. 2003;4:491–497. - PubMed
-
- Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nature Rev Cancer. 2006;6:776–788. - PubMed
-
- Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. - PubMed
-
- Cook WJ, Jeffrey LC, Sullivan ML, Vierstra RD. Three-dimensional structure of a ubiquitin-conjugating enzyme (E2) J Biol Chem. 1992;267:15116–15121. - PubMed
-
- Varadan R, et al. Solution conformation of Lys 63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

