Long-range chromosomal interactions and gene regulation
- PMID: 18931780
- PMCID: PMC2653627
- DOI: 10.1039/b803580f
Long-range chromosomal interactions and gene regulation
Abstract
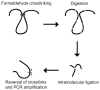
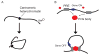
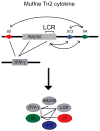
Over the last few years important new insights into the process of long-range gene regulation have been obtained. Gene regulatory elements are found to engage in direct physical interactions with distant target genes and with loci on other chromosomes to modulate transcription. An overview of recently discovered long-range chromosomal interactions is presented, and a network approach is proposed to unravel gene-element relationships. Gene expression is controlled by regulatory elements that can be located far away along the chromosome or in some cases even on other chromosomes. Genes and regulatory elements physically associate with each other resulting in complex genome-wide networks of chromosomal interactions. Here we describe several well-characterized cases of long-range interactions involved in the activation and repression of transcription. We speculate on how these interactions may affect gene expression and outline possible mechanisms that may facilitate encounters between distant elements. Finally, we propose that a genome-wide network analysis may provide new insights into the logic of long-range gene regulation.
Figures

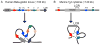
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

