ADAM function in embryogenesis
- PMID: 18935966
- PMCID: PMC2693894
- DOI: 10.1016/j.semcdb.2008.09.006
ADAM function in embryogenesis
Abstract
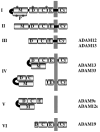
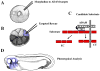
Cleavage of proteins inserted into the plasma membrane (shedding) is an essential process controlling many biological functions including cell signaling, cell adhesion and migration as well as proliferation and differentiation. ADAM surface metalloproteases have been shown to play an essential role in these processes. Gene inactivation during embryonic development have provided evidence of the central role of ADAM proteins in nematodes, flies, frogs, birds and mammals. The relative contribution of four subfamilies of ADAM proteins to developmental processes is the focus of this review.
Figures


References
-
- Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. - PubMed
-
- Muga A, Neugebauer W, Hirama T, Surewicz WK. Membrane interaction and conformational properties of the putative fusion peptide of PH-30, a protein active in sperm-egg fusion. Biochemistry. 1994;33:4444–4448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

