RNA folding and ribosome assembly
- PMID: 18935976
- PMCID: PMC2651837
- DOI: 10.1016/j.cbpa.2008.09.024
RNA folding and ribosome assembly
Abstract
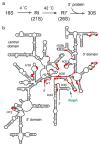
Ribosome synthesis is a tightly regulated process that is crucial for cell survival. Chemical footprinting, mass spectrometry, and cryo-electron microscopy are revealing how these complex cellular machines are assembled. Rapid folding of the rRNA provides a platform for protein-induced assembly of the bacterial 30S ribosome. Multiple assembly pathways increase the flexibility of the assembly process, while accessory factors and modification enzymes chaperone the late stages of assembly and control the quality of the mature subunits.
Figures


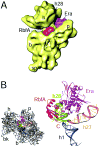
References
-
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. - PubMed
-
- Ramakrishnan V, Moore PB. Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol. 2001;11:144–154. - PubMed
-
- Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J Mol Biol. 1969;40:391–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

