Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry
- PMID: 18936057
- PMCID: PMC2649805
- DOI: 10.1074/mcp.M800232-MCP200
Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry
Abstract
We present results from a novel strategy that enables concurrent identification of protein-protein interactions and topologies in living cells without specific antibodies or genetic manipulations for immuno-/affinity purifications. The strategy consists of (i) a chemical cross-linking reaction: intact cell labeling with a novel class of chemical cross-linkers, protein interaction reporters (PIRs); (ii) two-stage mass spectrometric analysis: stage 1 identification of PIR-labeled proteins and construction of a restricted database by two-dimensional LC/MSMS and stage 2 analysis of PIR-labeled peptides by multiplexed LC/FTICR-MS; and (iii) data analysis: identification of cross-linked peptides and proteins of origin using accurate mass and other constraints. The primary advantage of the PIR approach and distinction from current technology is that protein interactions together with topologies are detected in native biological systems by stabilizing protein complexes with new covalent bonds while the proteins are present in the original cellular environment. Thus, weak or transient interactions or interactions that require properly folded, localized, or membrane-bound proteins can be labeled and identified through the PIR approach. This strategy was applied to Shewanella oneidensis bacterial cells, and initial studies resulted in identification of a set of protein-protein interactions and their contact/binding regions. Furthermore most identified interactions involved membrane proteins, suggesting that the PIR approach is particularly suited for studies of membrane protein-protein interactions, an area under-represented with current widely used approaches.
Figures


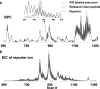
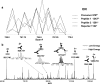

References
-
- Fields, S., and Song, O. ( 1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 - PubMed
-
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 - PubMed
-
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. ( 1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 - PubMed
-
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. ( 2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 - PubMed
-
- Zhu, H., and Snyder, M. ( 2003) Protein chip technology. Curr. Opin. Chem. Biol. 7, 55–63 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

