An enhanced mass spectrometry approach reveals human embryonic stem cell growth factors in culture
- PMID: 18936058
- PMCID: PMC2649806
- DOI: 10.1074/mcp.M800190-MCP200
An enhanced mass spectrometry approach reveals human embryonic stem cell growth factors in culture
Abstract
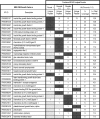
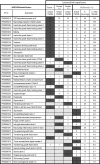
The derivation and long-term maintenance of human embryonic stem cells (hESCs) has been established in culture formats that are both dependent and independent of support (feeder) cells. However, the factors responsible for preserving the viability of hESCs in a nascent state remain unknown. We describe a mass spectrometry-based method for probing the secretome of the hESC culture microenvironment to identify potential regulating protein factors that are in low abundance. Individual samples were analyzed several times, using successive mass (m/z) and retention time-directed exclusion, without sampling the same peptide ion twice. This iterative exclusion -mass spectrometry (IE-MS) approach more than doubled protein and peptide metrics in comparison to a simple repeat analysis method on the same instrument, even after extensive sample pre-fractionation. Furthermore, implementation of the IE-MS approach was shown to enhance the performance of an older quadrupole time of flight (Q-ToF) MS. The resulting number of identified peptides approached that of a parallel repeat analysis on a newer LTQ-Orbitrap MS. The combination of the results of both instruments proved to be superior to that achieved by a single instrument in the identification of additional proteins. Using the IE-MS strategy, combined with complementary gel- and solution-based fractionation methods, the hESC culture microenvironment was extensively probed. Over 10 to 12 times more extracellular proteins were observed compared with previously published surveys. The detection of previously undetectable growth factors, present at concentrations ranging from 10(-9) to 10(-11) g/ml, highlights the depth of our profiling. The IE-MS approach provides a simple and reliable technique that greatly enhances instrument performance by increasing the effective depth of MS-based proteomic profiling. This approach should be widely applicable to any LC-MS/MS instrument platform or biological system.
Figures

References
-
- Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A., and Bongso, A. ( 2000) Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404 - PubMed
-
- Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. ( 1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 - PubMed
-
- Menendez, P., Wang, L., and Bhatia, M. ( 2005) Genetic manipulation of human embryonic stem cells: a system to study early human development and potential therapeutic applications. Curr. Gene Ther. 5, 375–385 - PubMed
-
- Evans, M. J., and Kaufman, M. H. ( 1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 - PubMed
-
- Hoffman, L. M., and Carpenter, M. K. ( 2005) Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 23, 699–708 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

