Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome
- PMID: 18936163
- PMCID: PMC2612491
- DOI: 10.1128/MCB.00997-08
Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome
Abstract
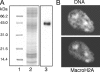
We studied the enrichment and distribution of the histone variant mH2A1 in the condensed inactive X (Xi) chromosome. By using highly specific antibodies against mH2A1 and stable HEK 293 cell lines expressing either green fluorescent protein (GFP)-mH2A1 or GFP-H2A, we found that the Xi chromosome contains approximately 1.5-fold more mH2A1 than the autosomes. To determine the in vivo distribution of mH2A1 along the X chromosome, we used a native chromatin immunoprecipitation-on-chip technique. DNA isolated from mH2A1-immunoprecipitated nucleosomes from either male or female mouse liver were hybridized to tiling microarrays covering 5 kb around most promoters or the entire X chromosome. The data show that mH2A1 is uniformly distributed across the entire Xi chromosome. Interestingly, a stronger mH2A1 enrichment along the pseudoautosomal X chromosome region was observed in both sexes. Our results indicate a potential role for macroH2A in large-scale chromosome structure and genome stability.
Figures




References
-
- Abbott, D. W., V. S. Ivanova, X. Wang, W. M. Bonner, and J. Ausio. 2001. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 27641945-41949. - PubMed
-
- Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant MacroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 111033-1041. - PubMed
-
- Ausió, J., and D. W. Abbott. 2002. The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 415945-5949. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
