The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis
- PMID: 18936164
- PMCID: PMC2612497
- DOI: 10.1128/MCB.00315-08
The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis
Abstract
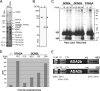
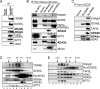
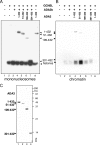
Human STAGA is a multisubunit transcriptional coactivator containing the histone acetyltransferase GCN5L. Previous studies of the related yeast SAGA complex have shown that the yeast Gcn5, Ada2, and Ada3 components form a heterotrimer that is important for the enzymatic function of SAGA. Here, we report that ADA2a and ADA2b, two human homologues of yeast Ada2, each have the ability to form a heterotrimer with ADA3 and GCN5L but that only the ADA2b homologue is found in STAGA. By comparing the patterns of acetylation of several substrates, we found context-dependent requirements for ADA2b and ADA3 for the efficient acetylation of histone tails by GCN5. With human proteins, unlike yeast proteins, the acetylation of free core histones by GCN5 is unaffected by ADA2b or ADA3. In contrast, the acetylation of mononucleosomal substrates by GCN5 is enhanced by ADA2b, with no significant additional effect of ADA3, and the efficient acetylation of nucleosomal arrays (chromatin) by GCN5 requires both ADA2b and ADA3. Thus, ADA2b and ADA3 appear to act at two different levels of histone organization within chromatin to facilitate GCN5 function. Interestingly, although ADA2a forms a complex(es) with GCN5 and ADA3 both in vitro and in vivo, ADA2a-containing complexes are unable to acetylate nucleosomal H3. We have also shown the preferential recruitment of ADA2b, relative to ADA2a, to p53-dependent genes. This finding indicates that the previously demonstrated presence and function of GCN5 on these promoters reflect the action of STAGA and that the ADA2a and ADA2b paralogues have nonredundant functional roles.
Figures





References
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. - PubMed
-
- Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2777989-7995. - PubMed
-
- Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 236944-6957. - PMC - PubMed
-
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 81243-1254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
