Sam68 regulates a set of alternatively spliced exons during neurogenesis
- PMID: 18936165
- PMCID: PMC2612485
- DOI: 10.1128/MCB.01349-08
Sam68 regulates a set of alternatively spliced exons during neurogenesis
Abstract
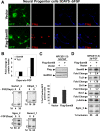
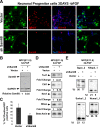
Sam68 (Src-associated in mitosis, 68 kDa) is a KH domain RNA binding protein implicated in a variety of cellular processes, including alternative pre-mRNA splicing, but its functions are not well understood. Using RNA interference knockdown of Sam68 expression and splicing-sensitive microarrays, we identified a set of alternative exons whose splicing depends on Sam68. Detailed analysis of one newly identified target exon in epsilon sarcoglycan (Sgce) showed that both RNA elements distributed across the adjacent introns and the RNA binding activity of Sam68 are necessary to repress the Sgce exon. Sam68 protein is upregulated upon neuronal differentiation of P19 cells, and many Sam68 RNA targets change in expression and splicing during this process. When Sam68 is knocked down by short hairpin RNAs, many Sam68-dependent splicing changes do not occur and P19 cells fail to differentiate. We also found that the differentiation of primary neuronal progenitor cells from embryonic mouse neocortex is suppressed by Sam68 depletion and promoted by Sam68 overexpression. Thus, Sam68 controls neurogenesis through its effects on a specific set of RNA targets.
Figures

References
-
- Ao, W., J. Gaudet, W. J. Kent, S. Muttumu, and S. E. Mango. 2004. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 3051743-1746. - PubMed
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 228-36. - PubMed
-
- Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 1322-29. - PubMed
-
- Baumann, C. A., V. Ribon, M. Kanzaki, D. C. Thurmond, S. Mora, S. Shigematsu, P. E. Bickel, J. E. Pessin, and A. R. Saltiel. 2000. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 407202-207. - PubMed
-
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
