Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods
- PMID: 18936483
- PMCID: PMC2579371
- DOI: 10.1073/pnas.0806548105
Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods
Abstract
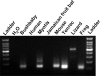
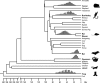
Horizontal transfer (HT) is central to the evolution of prokaryotic species. Selfish and mobile genetic elements, such as phages, plasmids, and transposons, are the primary vehicles for HT among prokaryotes. In multicellular eukaryotes, the prevalence and evolutionary significance of HT remain unclear. Here, we identified a set of DNA transposon families dubbed SPACE INVADERS (or SPIN) whose consensus sequences are approximately 96% identical over their entire length (2.9 kb) in the genomes of murine rodents (rat/mouse), bushbaby (prosimian primate), little brown bat (laurasiatherian), tenrec (afrotherian), opossum (marsupial), and two non-mammalian tetrapods (anole lizard and African clawed frog). In contrast, SPIN elements were undetectable in other species represented in the sequence databases, including 19 other mammals with draft whole-genome assemblies. This patchy distribution, coupled with the extreme level of SPIN identity in widely divergent tetrapods and the overall lack of selective constraint acting on these elements, is incompatible with vertical inheritance, but strongly indicative of multiple horizontal introductions. We show that these germline infiltrations likely occurred around the same evolutionary time (15-46 mya) and spawned some of the largest bursts of DNA transposon activity ever recorded in any species lineage (nearly 100,000 SPIN copies per haploid genome in tenrec). The process also led to the emergence of a new gene in the murine lineage derived from a SPIN transposase. In summary, HT of DNA transposons has contributed significantly to shaping and diversifying the genomes of multiple mammalian and tetrapod species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
A new SPIN on horizontal transfer.Proc Natl Acad Sci U S A. 2008 Nov 4;105(44):16827-8. doi: 10.1073/pnas.0809525105. Epub 2008 Oct 30. Proc Natl Acad Sci U S A. 2008. PMID: 18974220 Free PMC article. No abstract available.
References
-
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. - PubMed
-
- Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. - PubMed
-
- Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. - PubMed
-
- Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

