SSB as an organizer/mobilizer of genome maintenance complexes
- PMID: 18937104
- PMCID: PMC2583361
- DOI: 10.1080/10409230802341296
SSB as an organizer/mobilizer of genome maintenance complexes
Abstract
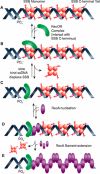
When duplex DNA is altered in almost any way (replicated, recombined, or repaired), single strands of DNA are usually intermediates, and single-stranded DNA binding (SSB) proteins are present. These proteins have often been described as inert, protective DNA coatings. Continuing research is demonstrating a far more complex role of SSB that includes the organization and/or mobilization of all aspects of DNA metabolism. Escherichia coli SSB is now known to interact with at least 14 other proteins that include key components of the elaborate systems involved in every aspect of DNA metabolism. Most, if not all, of these interactions are mediated by the amphipathic C-terminus of SSB. In this review, we summarize the extent of the eubacterial SSB interaction network, describe the energetics of interactions with SSB, and highlight the roles of SSB in the process of recombination. Similar themes to those highlighted in this review are evident in all biological systems.
Figures


References
-
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171(4361):964. - PubMed
-
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737. - PubMed
-
- Varghese AJ. Photochemistry of nucleic acids and their constituents. Photophysiology. 1972;(7):207. - PubMed
-
- Hanawalt PC. The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem Photobiol. 1966;5(1):1. - PubMed
-
- Hanawalt PC. Normal replication of DNA after repair replication in bacteria. Nature. 1967;214(5085):269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
