Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase
- PMID: 18937500
- PMCID: PMC2892342
- DOI: 10.1021/bi801604x
Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase
Abstract
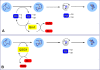
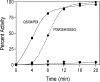
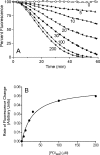
The flavin-dependent quiescin-sulfhydryl oxidase (QSOX) inserts disulfide bridges into unfolded reduced proteins with the reduction of molecular oxygen to form hydrogen peroxide. This work investigates how QSOX and protein disulfide isomerase (PDI) cooperate in vitro to generate native pairings in two unfolded reduced proteins: ribonuclease A (RNase, four disulfide bonds and 105 disulfide isomers of the fully oxidized protein) and avian riboflavin binding protein (RfBP, nine disulfide bonds and more than 34 million corresponding disulfide pairings). Experiments combining avian or human QSOX with up to 200 muM avian or human reduced PDI show that the isomerase is not a significant substrate of QSOX. Both reduced RNase and RfBP can be efficiently refolded in an aerobic solution containing micromolar concentrations of reduced PDI and nanomolar levels of QSOX without any added oxidized PDI or glutathione redox buffer. Refolding of RfBP is followed continuously using the complete quenching of the fluorescence of free riboflavin that occurs on binding to apo-RfBP. The rate of refolding is half-maximal at 30 muM reduced PDI when the reduced client protein (1 muM) is used in the presence of 30 nM QSOX. The use of high concentrations of PDI, in considerable excess over the folding protein client, reflects the concentration prevailing in the lumen of the endoplasmic reticulum and allows the redox poise of these in vitro experiments to be set with oxidized and reduced PDI. In the absence of either QSOX or redox buffer, the fastest refolding of RfBP is accomplished with excess reduced PDI and just enough oxidized PDI to generate nine disulfides in the protein client. These in vitro experiments are discussed in terms of current models for oxidative folding in the endoplasmic reticulum.
Figures




References
-
- Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplamic reticulum. Mol. Cell. 1998;1:171–182. - PubMed
-
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1998;1:161–170. - PubMed
-
- Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J. Biol. Chem. 1999;274:31759–31762. - PubMed
-
- Benayoun B, Esnard-Fève A, Castella S, Courty Y, Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase:biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J. Biol. Chem. 2001;276:13830–13837. - PubMed
-
- Dias-Gunasekara S, Gubbens J, van Lith M, Dunne C, Williams JA, Kataky R, Scoones D, Lapthorn A, Bulleid NJ, Benham AM. Tissue-specific expression and dimerization of the endoplasmic reticulum oxidoreductase Ero1beta. J. Biol. Chem. 2005;280:33066–33075. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

