Empirical Bayes accomodation of batch-effects in microarray data using identical replicate reference samples: application to RNA expression profiling of blood from Duchenne muscular dystrophy patients
- PMID: 18937867
- PMCID: PMC2576259
- DOI: 10.1186/1471-2164-9-494
Empirical Bayes accomodation of batch-effects in microarray data using identical replicate reference samples: application to RNA expression profiling of blood from Duchenne muscular dystrophy patients
Abstract
Background: Non-biological experimental error routinely occurs in microarray data collected in different batches. It is often impossible to compare groups of samples from independent experiments because batch effects confound true gene expression differences. Existing methods can correct for batch effects only when samples from all biological groups are represented in every batch.
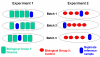
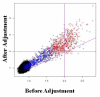
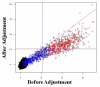
Results: In this report we describe a generalized empirical Bayes approach to correct for cross-experimental batch effects, allowing direct comparisons of gene expression between biological groups from independent experiments. The proposed experimental design uses identical reference samples in each batch in every experiment. These reference samples are from the same tissue as the experimental samples. This design with tissue matched reference samples allows a gene-by-gene correction to be performed using fewer arrays than currently available methods. We examine the effects of non-biological variation within a single experiment and between experiments.
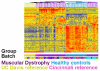
Conclusion: Batch correction has a significant impact on which genes are identified as differentially regulated. Using this method, gene expression in the blood of patients with Duchenne Muscular Dystrophy is shown to differ for hundreds of genes when compared to controls. The numbers of specific genes differ depending upon whether between experiment and/or between batch corrections are performed.
Figures



References
-
- Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, (eds) The analysis of gene expression data: methods and software. New York Springer; 2003.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

