In silico microarray probe design for diagnosis of multiple pathogens
- PMID: 18940003
- PMCID: PMC2596143
- DOI: 10.1186/1471-2164-9-496
In silico microarray probe design for diagnosis of multiple pathogens
Abstract
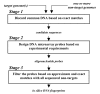
Background: With multiple strains of various pathogens being sequenced, it is necessary to develop high-throughput methods that can simultaneously process multiple bacterial or viral genomes to find common fingerprints as well as fingerprints that are unique to each individual genome. We present algorithmic enhancements to an existing single-genome pipeline that allows for efficient design of microarray probes common to groups of target genomes. The enhanced pipeline takes advantage of the similarities in the input genomes to narrow the search to short, nonredundant regions of the target genomes and, thereby, significantly reduces the computation time. The pipeline also computes a three-state hybridization matrix, which gives the expected hybridization of each probe with each target.
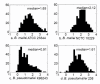
Results: Design of microarray probes for eight pathogenic Burkholderia genomes shows that the multiple-genome pipeline is nearly four-times faster than the single-genome pipeline for this application. The probes designed for these eight genomes were experimentally tested with one non-target and three target genomes. Hybridization experiments show that less than 10% of the designed probes cross hybridize with non-targets. Also, more than 65% of the probes designed to identify all Burkholderia mallei and B. pseudomallei strains successfully hybridize with a B. pseudomallei strain not used for probe design.
Conclusion: The savings in runtime suggest that the enhanced pipeline can be used to design fingerprints for tens or even hundreds of related genomes in a single run. Hybridization results with an unsequenced B. pseudomallei strain indicate that the designed probes might be useful in identifying unsequenced strains of B. mallei and B. pseudomallei.
Figures
Similar articles
-
Genomic fingerprinting of Burkholderia pseudomallei and B. mallei pathogens with DNA array based on interspecies sequence differences obtained by subtractive hybridization.Res Microbiol. 2006 Sep;157(7):684-92. doi: 10.1016/j.resmic.2006.02.008. Epub 2006 Mar 27. Res Microbiol. 2006. PMID: 16621455
-
DNA microarray-based detection and identification of Burkholderia mallei, Burkholderia pseudomallei and Burkholderia spp.Mol Cell Probes. 2009 Jun-Aug;23(3-4):178-87. doi: 10.1016/j.mcp.2009.04.001. Epub 2009 Apr 12. Mol Cell Probes. 2009. PMID: 19366627
-
Development of a polymerase chain reaction assay for the specific identification of Burkholderia mallei and differentiation from Burkholderia pseudomallei and other closely related Burkholderiaceae.Diagn Microbiol Infect Dis. 2006 May;55(1):37-45. doi: 10.1016/j.diagmicrobio.2005.11.007. Epub 2006 Mar 20. Diagn Microbiol Infect Dis. 2006. PMID: 16546342
-
DNA microarray technology: a new tool for the epidemiological typing of bacterial pathogens?FEMS Immunol Med Microbiol. 2006 Jul;47(2):178-89. doi: 10.1111/j.1574-695X.2006.00081.x. FEMS Immunol Med Microbiol. 2006. PMID: 16831204 Review.
-
In-depth query of large genomes using tiling arrays.Methods Mol Biol. 2007;377:163-74. doi: 10.1007/978-1-59745-390-5_10. Methods Mol Biol. 2007. PMID: 17634616 Review.
Cited by
-
Alignment-free design of highly discriminatory diagnostic primer sets for Escherichia coli O104:H4 outbreak strains.PLoS One. 2012;7(4):e34498. doi: 10.1371/journal.pone.0034498. Epub 2012 Apr 5. PLoS One. 2012. PMID: 22496820 Free PMC article.
-
An algorithm of discovering signatures from DNA databases on a computer cluster.BMC Bioinformatics. 2014 Oct 5;15(1):339. doi: 10.1186/1471-2105-15-339. BMC Bioinformatics. 2014. PMID: 25282047 Free PMC article.
-
HTSFinder: Powerful Pipeline of DNA Signature Discovery by Parallel and Distributed Computing.Evol Bioinform Online. 2016 Feb 10;12:73-85. doi: 10.4137/EBO.S35545. eCollection 2016. Evol Bioinform Online. 2016. PMID: 26884678 Free PMC article.
-
Animal Viruses Probe dataset (AVPDS) for microarray-based diagnosis and identification of viruses.Curr Microbiol. 2014 Mar;68(3):301-4. doi: 10.1007/s00284-013-0477-4. Epub 2013 Oct 16. Curr Microbiol. 2014. PMID: 24129840 Free PMC article.
-
SNIT: SNP identification for strain typing.Source Code Biol Med. 2011 Sep 8;6:14. doi: 10.1186/1751-0473-6-14. Source Code Biol Med. 2011. PMID: 21902825 Free PMC article.
References
-
- Fitch JP, Gardner SN, Kuczmarski TA, Kurtz S, Myers R, Ott LL, Slezak TR, Vitalis EA, Zemla AT, McCready PM. Rapid Development of Nucleic Acid Diagnostics. Proceedings of the IEEE. 2002;90:1708–1720. doi: 10.1109/JPROC.2002.804680. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

