Structure of Seneca Valley Virus-001: an oncolytic picornavirus representing a new genus
- PMID: 18940610
- PMCID: PMC2572565
- DOI: 10.1016/j.str.2008.07.013
Structure of Seneca Valley Virus-001: an oncolytic picornavirus representing a new genus
Abstract
The crystal structure of Seneca Valley Virus-001 (SVV-001), the representative member of a new genus, Senecavirus, is reported at 2.3A resolution. SVV-001 is the first naturally occurring nonpathogenic picornavirus shown to mediate selective cytotoxicity towards tumor cells with neuroendocrine cancer features. The nonsegmented (+) ssRNA genome of SVV-001 shares closest sequence similarity with the genomes of the members of Cardiovirus. The overall tertiary structure of VP1-VP4 subunits is conserved with the exception of loops, especially those of VP1 that show large deviations relative to the members of the cardioviruses. The surface loops of VP1 and VP2 are predicted to mediate cell tropism of SVV-001. In addition, the organization of the packaged nucleic acid density indicates that certain regions of VP2 and VP4 interact closely with the packaged nucleic acid.
Figures
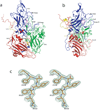
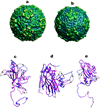


Similar articles
-
Crystallization and preliminary X-ray diffraction studies of Seneca Valley virus-001, a new member of the Picornaviridae family.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Apr 1;64(Pt 4):293-6. doi: 10.1107/S1744309108006921. Epub 2008 Mar 21. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18391430 Free PMC article.
-
Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus.J Gen Virol. 2008 May;89(Pt 5):1265-1275. doi: 10.1099/vir.0.83570-0. J Gen Virol. 2008. PMID: 18420805
-
Oncolytic Seneca Valley Virus: past perspectives and future directions.Oncolytic Virother. 2016 Sep 6;5:81-9. doi: 10.2147/OV.S96915. eCollection 2016. Oncolytic Virother. 2016. PMID: 27660749 Free PMC article. Review.
-
Characterisation of a Seneca Valley virus thermostable mutant.Virology. 2022 Oct;575:74-82. doi: 10.1016/j.virol.2022.08.015. Epub 2022 Aug 30. Virology. 2022. PMID: 36084546
-
Senecavirus A.Vet Pathol. 2017 Jan;54(1):11-21. doi: 10.1177/0300985816653990. Epub 2016 Jul 11. Vet Pathol. 2017. PMID: 27371541 Review.
Cited by
-
Immunogenicity of an Inactivated Senecavirus A Vaccine with a Contemporary Brazilian Strain in Mice.Vaccines (Basel). 2024 Jul 26;12(8):845. doi: 10.3390/vaccines12080845. Vaccines (Basel). 2024. PMID: 39203971 Free PMC article.
-
Isolation and Characterization of Seneca Valley Virus From Pig Transboundary Spread to the Mink Infection.Transbound Emerg Dis. 2025 Jun 16;2025:4428550. doi: 10.1155/tbed/4428550. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40551879 Free PMC article.
-
Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer.J Natl Cancer Inst. 2013 Jul 17;105(14):1059-65. doi: 10.1093/jnci/djt130. Epub 2013 Jun 5. J Natl Cancer Inst. 2013. PMID: 23739064 Free PMC article.
-
Senecavirus a 3D Interacts with NLRP3 to Induce IL-1β Production by Activating NF-κB and Ion Channel Signals.Microbiol Spectr. 2022 Apr 27;10(2):e0209721. doi: 10.1128/spectrum.02097-21. Epub 2022 Mar 7. Microbiol Spectr. 2022. PMID: 35254168 Free PMC article.
-
Non-human viruses developed as therapeutic agent for use in humans.Rev Med Virol. 2011 Jul;21(4):227-39. doi: 10.1002/rmv.694. Epub 2011 May 11. Rev Med Virol. 2011. PMID: 21560181 Free PMC article. Review.
References
-
- Au GG, Lindberg AM, Barry RD, Shafren DR. Oncolysis of vascular malignant human melanoma tumors by Coxsackievirus A21. International journal of oncology. 2005;26:1471–1476. - PubMed
-
- Bajaj C, Djeu P, Thane A, Siddavanahalli V. Interactive visual exploration of large flexible multi-component molecular complexes. Proc. of the Annual IEEEE Visualization Conference. 2004:243–250.
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;D54:905–921. - PubMed
-
- CCP4. The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;D50:760–763. - PubMed
-
- Cohen GE. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. Journal of Applied Crystallography. 1997;30:1160–1161.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

