A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding
- PMID: 18940669
- PMCID: PMC2677525
- DOI: 10.1016/j.chembiol.2008.09.008
A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding
Abstract
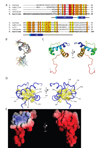
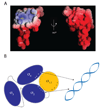
The sigma factors are the key regulators of bacterial transcription initiation. Through direct read-out of promoter DNA sequence, they recruit the core RNA polymerase to sites of initiation, thereby dictating the RNA polymerase promoter-specificity. The group 1 sigma factors, which direct the vast majority of transcription initiation during log phase growth and are essential for viability, are autoregulated by an N-terminal sequence known as sigma1.1. We report the solution structure of Thermotoga maritima sigmaA sigma1.1. We additionally demonstrate by using chemical crosslinking strategies that sigma1.1 is in close proximity to the promoter recognition domains of sigmaA. We therefore propose that sigma1.1 autoinhibits promoter DNA binding of free sigmaA by stabilizing a compact organization of the sigma factor domains that is unable to bind DNA.
Figures




References
-
- Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 2003;6:93–100. - PubMed
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. - PubMed
-
- Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

