Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance
- PMID: 18940787
- PMCID: PMC2600957
- DOI: 10.1534/genetics.108.095034
Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance
Abstract
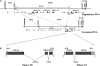
The rice blast resistance gene Pikm was cloned by a map-based cloning strategy. High-resolution genetic mapping and sequencing of the gene region in the Pikm-containing cultivar Tsuyuake narrowed down the candidate region to a 131-kb genomic interval. Sequence analysis predicted two adjacently arranged resistance-like genes, Pikm1-TS and Pikm2-TS, within this candidate region. These genes encoded proteins with a nucleotide-binding site (NBS) and leucine-rich repeats (LRRs) and were considered the most probable candidates for Pikm. However, genetic complementation analysis of transgenic lines individually carrying these two genes negated the possibility that either Pikm1-TS or Pikm2-TS alone was Pikm. Instead, it was revealed that transgenic lines carrying both of these genes expressed blast resistance. The results of the complementation analysis and an evaluation of the resistance specificity of the transgenic lines to blast isolates demonstrated that Pikm-specific resistance is conferred by cooperation of Pikm1-TS and Pikm2-TS. Although these two genes are not homologous with each other, they both contain all the conserved motifs necessary for an NBS-LRR class gene to function independently as a resistance gene.
Figures



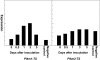
References
-
- Chen, X., J. Shang, D. Chen, C. Lei, Y. Zou et al., 2006. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46 794–804. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

