High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA
- PMID: 18940866
- PMCID: PMC2588502
- DOI: 10.1093/nar/gkn725
High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA
Abstract
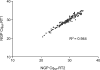
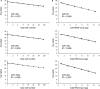
MicroRNAs (miRNAs) are an emerging class of small non-coding RNAs implicated in a wide variety of cellular processes. Research in this field is accelerating, and the growing number of miRNAs emphasizes the need for high-throughput and sensitive detection methods. Here we present the successful evaluation of the Megaplex reverse transcription format of the stem-loop primer-based real-time quantitative polymerase chain reaction (RT-qPCR) approach to quantify miRNA expression. The Megaplex reaction provides simultaneous reverse transcription of 450 mature miRNAs, ensuring high-throughput detection. Further, the introduction of a complementary DNA pre-amplification step significantly reduces the amount of input RNA needed, even down to single-cell level. To evaluate possible pre-amplification bias, we compared the expression of 384 miRNAs in three different cancer cell lines with Megaplex RT, with or without an additional pre-amplification step. The normalized Cq values of all three sample pairs showed a good correlation with maintenance of differential miRNA expression between the cell lines. Moreover, pre-amplification using 10 ng of input RNA enabled the detection of miRNAs that were undetectable when using Megaplex alone with 400 ng of input RNA. The high specificity of RT-qPCR together with a superior sensitivity makes this approach the method of choice for high-throughput miRNA expression profiling.
Figures
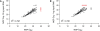
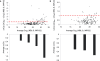
References
-
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. - PubMed
-
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
-
- Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

