MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes
- PMID: 18940874
- PMCID: PMC2608843
- DOI: 10.1093/dnares/dsn027
MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes
Abstract
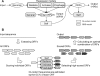
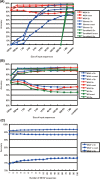
Recent advances in DNA sequencers are accelerating genome sequencing, especially in microbes, and complete and draft genomes from various species have been sequenced in rapid succession. Here, we present a comprehensive gene prediction tool, the MetaGeneAnnotator (MGA), which precisely predicts all kinds of prokaryotic genes from a single or a set of anonymous genomic sequences having a variety of lengths. The MGA integrates statistical models of prophage genes, in addition to those of bacterial and archaeal genes, and also uses a self-training model from input sequences for predictions. As a result, the MGA sensitively detects not only typical genes but also atypical genes, such as horizontally transferred and prophage genes in a prokaryotic genome. In this paper, we also propose a novel approach for analyzing the ribosomal binding site (RBS), which enables us to detect species-specific patterns of the RBSs. The MGA has the ingenious RBS model based on this approach, and precisely predicts translation starts of genes. The MGA also succeeds in improving prediction accuracies for short sequences by using the adapted RBS models (96% sensitivity and 93% specificity for 700 bp fragments). These features of the MGA expedite wide ranges of microbial genome studies, such as genome annotations and metagenome analyses.
Figures
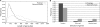



References
-
- Borodovsky M. Y., Sprizhitskii Y. A., Golovanov E. I., Aleksandrov A. A. Statistical patterns in primary structures of functional regions in the E. coli genome: III. Computer recognition of coding regions. Mol. Biol. 1986;20:1145–1150.
-
- Borodovsky M. Y., McIninch J. D. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 1993;17:123–153.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

