Impact of orthologous gene replacement on the circuitry governing pilus gene transcription in streptococci
- PMID: 18941636
- PMCID: PMC2565503
- DOI: 10.1371/journal.pone.0003450
Impact of orthologous gene replacement on the circuitry governing pilus gene transcription in streptococci
Abstract
Background: The evolutionary history of several genes of the bacterial pathogen Streptococcus pyogenes strongly suggests an origin in another species, acquired via replacement of the counterpart gene (ortholog) following a recombination event. An example of orthologous gene replacement is provided by the nra/rofA locus, which encodes a key regulator of pilus gene transcription. Of biological importance is the previous finding that the presence of the nra- and rofA-lineage alleles, which are approximately 35% divergent, correlates strongly with genetic markers for streptococcal infection at different tissue sites in the human host (skin, throat).
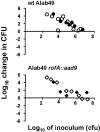
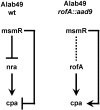
Methodology/principal findings: In this report, the impact of orthologous gene replacement targeting the nra/rofA locus is experimentally addressed. Replacement of the native nra-lineage allele with a rofA-lineage allele, plus their respective upstream regions, preserved the polarity of Nra effects on pilus gene transcription (i.e., activation) in the skin strain Alab49. Increased pilus gene transcription in the rofA chimera correlated with a higher rate of bacterial growth at the skin. The transcriptional regulator MsmR, which represses nra and pilus gene transcription in the Alab49 parent strain, has a slight activating effect on pilus gene expression in the rofA chimera construct.
Conclusions/significance: Data show that exchange of orthologous forms of a regulatory gene is stable and robust, and pathogenicity is preserved. Yet, new phenotypes may also be introduced by altering the circuitry within a complex transcriptional regulatory network. It is proposed that orthologous gene replacement via interspecies exchange is an important mechanism in the evolution of highly recombining bacteria such as S. pyogenes.
Conflict of interest statement
Figures







Similar articles
-
Heterogeneity in the polarity of Nra regulatory effects on streptococcal pilus gene transcription and virulence.Infect Immun. 2008 Jun;76(6):2490-7. doi: 10.1128/IAI.01567-07. Epub 2008 Mar 17. Infect Immun. 2008. PMID: 18347035 Free PMC article.
-
Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes.J Bacteriol. 2005 Jun;187(12):4163-72. doi: 10.1128/JB.187.12.4163-4172.2005. J Bacteriol. 2005. PMID: 15937178 Free PMC article.
-
The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region.Infect Immun. 2007 Dec;75(12):5698-710. doi: 10.1128/IAI.00175-07. Epub 2007 Sep 24. Infect Immun. 2007. PMID: 17893125 Free PMC article.
-
Thermosensitive pilus production by FCT type 3 Streptococcus pyogenes controlled by Nra regulator translational efficiency.Mol Microbiol. 2020 Jan;113(1):173-189. doi: 10.1111/mmi.14408. Epub 2019 Nov 11. Mol Microbiol. 2020. PMID: 31633834 Free PMC article.
-
Characterization of M-Type-Specific Pilus Expression in Group A Streptococcus.J Bacteriol. 2022 Nov 15;204(11):e0027022. doi: 10.1128/jb.00270-22. Epub 2022 Oct 26. J Bacteriol. 2022. PMID: 36286511 Free PMC article.
Cited by
-
Incremental Contributions of FbaA and Other Impetigo-Associated Surface Proteins to Fitness and Virulence of a Classical Group A Streptococcal Skin Strain.Infect Immun. 2017 Oct 18;85(11):e00374-17. doi: 10.1128/IAI.00374-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28808160 Free PMC article.
-
Natural disruption of two regulatory networks in serotype M3 group A Streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype.Infect Immun. 2014 May;82(5):1744-54. doi: 10.1128/IAI.01639-13. Epub 2014 Feb 10. Infect Immun. 2014. PMID: 24516115 Free PMC article.
-
Differences in SpeB protease activity among group A streptococci associated with superficial, invasive, and autoimmune disease.PLoS One. 2017 May 17;12(5):e0177784. doi: 10.1371/journal.pone.0177784. eCollection 2017. PLoS One. 2017. PMID: 28545045 Free PMC article.
-
Tissue tropisms in group A Streptococcus: what virulence factors distinguish pharyngitis from impetigo strains?Curr Opin Infect Dis. 2016 Jun;29(3):295-303. doi: 10.1097/QCO.0000000000000262. Curr Opin Infect Dis. 2016. PMID: 26895573 Free PMC article. Review.
-
Evolution of transcriptional regulatory circuits in bacteria.Cell. 2009 Jul 23;138(2):233-44. doi: 10.1016/j.cell.2009.07.002. Cell. 2009. PMID: 19632175 Free PMC article.
References
-
- Hanage WP, Fraser C, Spratt BG. The impact of homologous recombination on the generation of diversity in bacteria. Journal of Theoretical Biology. 2006;239:210–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

