Insertional gene activation by lentiviral and gammaretroviral vectors
- PMID: 18945765
- PMCID: PMC2612344
- DOI: 10.1128/JVI.01865-08
Insertional gene activation by lentiviral and gammaretroviral vectors
Abstract
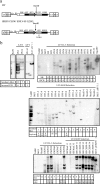
Gammaretroviral and lentiviral vectors are promising tools for gene therapy, but they can be oncogenic. The development of safer vectors depends on a quantitative assay for insertional mutagenesis. Here we report a rapid, inexpensive, and reproducible assay which uses a murine cell line to measure the frequency of interleukin-3 (IL-3)-independent mutants. Lentiviral and gammaretroviral vectors cause insertional mutagenesis at similar frequencies; however, they use different mechanisms. Human immunodeficiency virus (HIV)-based vectors generate mutants by insertion only into the growth hormone receptor (Ghr) locus. The HIV enhancer/promoter is active in the absence of the HIV Tat protein in this locus, and an HIV/Ghr spliced transcript expresses GHR and cells respond to GH. Deletion of the enhancer/promoter in a self-inactivating HIV-based vector prevents this mechanism of insertional mutagenesis. In contrast, gammaretroviral vectors insert into other loci, including IL-3 and genes identified as common insertion sites in the Retroviral Tagged Cancer Gene Database (RTCGD).
Figures



References
-
- Aiuti, A., S. Slavin, M. Aker, F. Ficara, S. Deola, A. Mortellaro, S. Morecki, G. Andolfi, A. Tabucchi, F. Carlucci, E. Marinello, F. Cattaneo, S. Vai, P. Servida, R. Miniero, M. G. Roncarolo, and C. Bordignon. 2002. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 2962410-2413. - PubMed
-
- Baixeras, E., S. Jeay, P. A. Kelly, and M. C. Postel-Vinay. 2001. The proliferative and antiapoptotic actions of growth hormone and insulin-like growth factor-1 are mediated through distinct signaling pathways in the Pro-B Ba/F3 cell line. Endocrinology 1422968-2977. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

