Afferent deprivation elicits a transcriptional response associated with neuronal survival after a critical period in the mouse cochlear nucleus
- PMID: 18945907
- PMCID: PMC2585504
- DOI: 10.1523/JNEUROSCI.2697-08.2008
Afferent deprivation elicits a transcriptional response associated with neuronal survival after a critical period in the mouse cochlear nucleus
Abstract
The mechanisms underlying enhanced plasticity of synaptic connections and susceptibilities to manipulations of afferent activity in developing sensory systems are not well understood. One example is the rapid and dramatic neuron death that occurs after removal of afferent input to the cochlear nucleus (CN) of young mammals and birds. The molecular basis of this critical period of neuronal vulnerability and the transition to survival independent of afferent input remains to be defined. Here we used microarray analyses, real-time reverse transcription PCR, and immunohistochemistry of the mouse CN to show that deafferentation results in strikingly different sets of regulated genes in vulnerable [postnatal day (P)7] and invulnerable (P21) CN. An unexpectedly large set of immune-related genes was induced by afferent deprivation after the critical period, which corresponded with glial proliferation over the same time frame. Apoptotic gene expression was not highly regulated in the vulnerable CN after afferent deprivation but, surprisingly, did increase after deafferentation at P21, when all neurons ultimately survive. Pharmacological activity blockade in the eighth nerve mimicked afferent deprivation for only a subset of the afferent deprivation regulated genes, indicating the presence of an additional factor not dependent on action potential-mediated signaling that is also responsible for transcriptional changes. Overall, our results suggest that the cell death machinery during this critical period is mainly constitutive, whereas after the critical period neuronal survival could be actively promoted by both constitutive and induced gene expression.
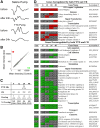
Figures







References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Bains JS, Shaw CA. Neurodegenerative disorders in humans: the role of glutathione in oxidative stress-mediated neuronal death. Brain Res Brain Res Rev. 1997;25:335–358. - PubMed
-
- Benn SC, Perrelet D, Kato AC, Scholz J, Decosterd I, Mannion RJ, Bakowska JC, Woolf CJ. Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron. 2002;36:45–56. - PubMed
-
- Boise LH, González-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
