RNA-dependent recruitment of the origin recognition complex
- PMID: 18946490
- PMCID: PMC2585170
- DOI: 10.1038/emboj.2008.221
RNA-dependent recruitment of the origin recognition complex
Abstract
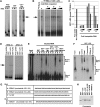
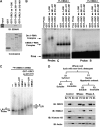
The origin recognition complex (ORC) has an important function in determining the initiation sites of DNA replication. In higher eukaryotes, ORC lacks sequence-specific DNA binding, and the mechanisms of ORC recruitment and origin determination are poorly understood. ORC is recruited with high efficiency to the Epstein-Barr virus origin of plasmid replication (OriP) through a complex mechanism involving interactions with the virus-encoded EBNA1 protein. We present evidence that ORC recruitment to OriP and DNA replication function depends on RGG-like motifs, referred to as LR1 and LR2, in the EBNA1 amino-terminal domain. Moreover, we show that LR1 and LR2 recruitment of ORC is RNA dependent. HMGA1a, which can functionally substitute for LR1 and LR2 domain, can also recruit ORC in an RNA-dependent manner. EBNA1 and HMGA1a RGG motifs bound to structured G-rich RNA, as did ORC1 peptides, which interact with EBNA1. RNase A treatment of cellular chromatin released a fraction of the total ORC, suggesting that ORC association with chromatin, and possibly cellular origins, is stabilized by RNA. We propose that structural RNA molecules mediate ORC recruitment at some cellular and viral origins, similar to OriP.
Figures





References
-
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023–2026 - PubMed
-
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 - PubMed
-
- Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16: 659–672 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

