Noncovalent probes for the investigation of structure and dynamics of protein-nucleic acid assemblies: the case of NC-mediated dimerization of genomic RNA in HIV-1
- PMID: 18946871
- PMCID: PMC2776628
- DOI: 10.1002/bip.21107
Noncovalent probes for the investigation of structure and dynamics of protein-nucleic acid assemblies: the case of NC-mediated dimerization of genomic RNA in HIV-1
Abstract
The nature of specific RNA-RNA and protein-RNA interactions involved in the process of genome dimerization and isomerization in HIV-1, which is mediated in vitro by stemloop 1 (SL1) of the packaging signal and by the nucleocapsid (NC) domain of the viral Gag polyprotein, was investigated by using archetypical nucleic acid ligands as noncovalent probes. Small-molecule ligands make contact with their target substrates through complex combinations of H-bonds, salt bridges, and hydrophobic interactions. Therefore, their binding patterns assessed by electrospray ionization mass spectrometry can provide valuable insights into the factors determining specific recognition between species involved in biopolymer assemblies. In the case of SL1, dimerization and isomerization create unique structural features capable of sustaining stable interactions with classic nucleic acid ligands. The binding modes exhibited by intercalators and minor groove binders were adversely affected by the significant distortion of the duplex formed by palindrome annealing in the kissing-loop (KL) dimer, whereas the modes observed for the corresponding extended duplex (ED) confirmed a more regular helical structure. Consistent with the ability to establish electrostatic interactions with highly negative pockets typical of helix anomalies, polycationic aminoglycosides bound to the stem-bulge motif conserved in all SL1 conformers, to the unpaired nucleotides located at the hinge between kissing hairpins in KL, and to the exposed bases flanking the palindrome duplex in ED. The patterns afforded by intercalators and minor groove binders did not display detectable variations when the corresponding NC-SL1 complexes were submitted to probing. In contrast, aminoglycosides displayed the ability to compete with the protein for overlapping sites, producing opposite effects on the isomerization process. Indeed, displacing NC from the stem-bulges of the KL dimer induced inhibition of stem melting and decreased the efficiency of isomerization. Competition for the hinge region, instead, eliminated the NC stabilization of a grip motif formed by nucleobases of opposite strands, thus facilitating the strand-exchange required for isomerization. These noncovalent probes provided further evidence that the structural context of the actual binding sites has significant influence on the chaperone activities of NC, which should be taken in account when developing potential drug candidates aimed at disrupting genome dimerization and isomerization in HIV-1.
2008 Wiley Periodicals, Inc.
Figures
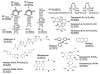
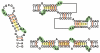

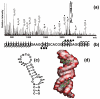

 symbol.
symbol.

References
-
- Walker TA, et al. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982;21:2320–29. - PubMed
-
- Krol A, Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–27. - PubMed
-
- Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

