A comparative analysis of viral matrix proteins using disorder predictors
- PMID: 18947403
- PMCID: PMC2579295
- DOI: 10.1186/1743-422X-5-126
A comparative analysis of viral matrix proteins using disorder predictors
Abstract
Background: A previous study (Goh G.K.-M., Dunker A.K., Uversky V.N. (2008) Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics. 9 (Suppl. 2), S4) revealed that HIV matrix protein p17 possesses especially high levels of predicted intrinsic disorder (PID). In this study, we analyzed the PID patterns in matrix proteins of viruses related and unrelated to HIV-1.
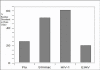
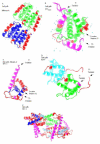
Results: Both SIVmac and HIV-1 p17 proteins were predicted by PONDR VLXT to be highly disordered with subtle differences containing 50% and 60% disordered residues, respectively. SIVmac is very closely related to HIV-2. A specific region that is predicted to be disordered in HIV-1 is missing in SIVmac. The distributions of PID patterns seem to differ in SIVmac and HIV-1 p17 proteins. A high level of PID for the matrix does not seem to be mandatory for retroviruses, since Equine Infectious Anemia Virus (EIAV), an HIV cousin, has been predicted to have low PID level for the matrix; i.e. its matrix protein p15 contains only 21% PID residues. Surprisingly, the PID percentage and the pattern of predicted disorder distribution for p15 resemble those of the influenza matrix protein M1 (25%).
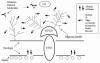
Conclusion: Our data might have important implications in the search for HIV vaccines since disorder in the matrix protein might provide a mechanism for immune evasion.
Figures

References
-
- Harris A, Sha B, Luo M. Structural similarities between influenza virus matrix protein M1 and human immunodeficiency virus matrix and capsid proteins: an evolutionary link between negative-stranded RNA viruses and retroviruses. J Gen Virol. 1999;80:863–869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

