Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components
- PMID: 18947425
- PMCID: PMC2613156
- DOI: 10.1186/1471-2148-8-290
Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components
Abstract
Background: Members of the eukaryote/archaea specific eRF1 and eRF3 protein families have central roles in translation termination. They are also central to various mRNA surveillance mechanisms, together with the eRF1 paralogue Dom34p and the eRF3 paralogues Hbs1p and Ski7p. We have examined the evolution of eRF1 and eRF3 families using sequence similarity searching, multiple sequence alignment and phylogenetic analysis.
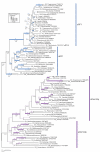
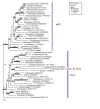
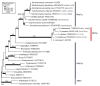
Results: Extensive BLAST searches confirm that Hbs1p and eRF3 are limited to eukaryotes, while Dom34p and eRF1 (a/eRF1) are universal in eukaryotes and archaea. Ski7p appears to be restricted to a subset of Saccharomyces species. Alignments show that Dom34p does not possess the characteristic class-1 RF minidomains GGQ, NIKS and YXCXXXF, in line with recent crystallographic analysis of Dom34p. Phylogenetic trees of the protein families allow us to reconstruct the evolution of mRNA surveillance mechanisms mediated by these proteins in eukaryotes and archaea.
Conclusion: We propose that the last common ancestor of eukaryotes and archaea possessed Dom34p-mediated no-go decay (NGD). This ancestral Dom34p may or may not have required a trGTPase, mostly like a/eEF1A, for its delivery to the ribosome. At an early stage in eukaryotic evolution, eEF1A was duplicated, giving rise to eRF3, which was recruited for translation termination, interacting with eRF1. eRF3 evolved nonsense-mediated decay (NMD) activity either before or after it was again duplicated, giving rise to Hbs1p, which we propose was recruited to assist eDom34p in eukaryotic NGD. Finally, a third duplication within ascomycete yeast gave rise to Ski7p, which may have become specialised for a subset of existing Hbs1p functions in non-stop decay (NSD). We suggest Ski7p-mediated NSD may be a specialised mechanism for counteracting the effects of increased stop codon read-through caused by prion-domain [PSI+] mediated eRF3 precipitation.
Figures

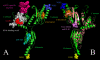

References
-
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M. et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. - PubMed
-
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. - PubMed
-
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. - PubMed
-
- Kobayashi T, Funakoshi Y, Hoshino S, Katada T. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J Biol Chem. 2004;279:45693–45700. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

