Aptamers that recognize drug-resistant HIV-1 reverse transcriptase
- PMID: 18948292
- PMCID: PMC2588506
- DOI: 10.1093/nar/gkn775
Aptamers that recognize drug-resistant HIV-1 reverse transcriptase
Abstract
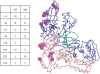
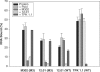
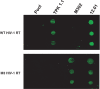
Drug-resistant variants of HIV-1 reverse transcriptase (RT) are also known to be resistant to anti-RT RNA aptamers. In order to be able to develop diagnostics and therapies that can focus on otherwise drug-resistant viruses, we have isolated two aptamers against a well-known, drug-resistant HIV-1 RT, Mutant 3 (M3) from the multidrug-resistant HIV-1 RT panel. One aptamer, M302, bound M3 but showed no significant affinity for wild-type (WT) HIV-1 RT, while another aptamer, 12.01, bound to both M3 and WT HIV-1 RTs. In contrast to all previously selected anti-RT aptamers, neither of these aptamers showed observable inhibition of either polymerase or RNase H activities. Aptamers M302 and 12.01 competed with one another for binding to M3, but they did not compete with a pseudoknot aptamer for binding to the template/primer cleft of WT HIV-1 RT. These results represent the surprising identification of an additional RNA-binding epitope on the surface of HIV-1 RT. M3 and WT HIV-1 RTs could be distinguished using an aptamer-based microarray. By probing protein conformation as a correlate to drug resistance we introduce an additional and useful measure for determining HIV-1 drug resistance.
Figures


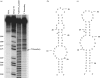



References
-
- Piacenti FJ. An update and review of antiretroviral therapy. Pharmacotherapy. 2006;26:1111–1133. - PubMed
-
- Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: spring 2008. Top. HIV Med. 2008;16:62–68. - PubMed
-
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. - PubMed
-
- Hirsch MS, Günthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, Hammer SM, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 2008;47:266–285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

