Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen
- PMID: 18948595
- PMCID: PMC2575446
- DOI: 10.1073/pnas.0806574105
Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen
Abstract
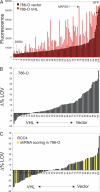
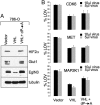
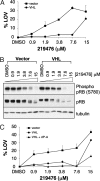
Clear cell renal carcinomas are the most common form of kidney cancer and frequently are linked to biallelic inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene. The VHL gene product, pVHL, has multiple functions including directing the polyubiquitylation of the HIF transcription factor. We screened 100 shRNA vectors, directed against 88 kinases, for their ability to inhibit the viability of VHL-/- renal carcinoma cells preferentially compared with isogenic cells in which pVHL function was restored. shRNAs for "hits" identified in the primary screen were interrogated in secondary screens that included shRNA titration studies. Multiple shRNAs against CDK6, MET, and MAP2K1 (also known as MEK1) preferentially inhibited the viability of 786-O and RCC4 VHL-/- cells compared with their wild-type pVHL-reconstituted counterparts. The sensitivity of pVHL-proficient cells to these shRNAs was not restored upon HIF activation, suggesting that loss of an hypoxia-inducible factor (HIF)-independent pVHL function formed the basis for selectivity. A small-molecule Cdk4/6 inhibitor displayed enhanced activity against VHL-/- renal carcinoma cells, suggesting that in some cases hits from shRNA screens such as described here might translate into therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hartwell L, Szankasi P, Roberts C, Murray A, Friend S. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. - PubMed
-
- Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. - PubMed
-
- Kaelin WG., Jr Cell biology: Divining cancer cell weaknesses. Nature. 2006;441:32–34. - PubMed
-
- Sharom JR, Bellows DS, Tyers M. From large networks to small molecules. Curr Opin Chem Biol. 2004;8:81–90. - PubMed
-
- Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

