Mutation in EGFP domain of LDL receptor-related protein 6 impairs cellular LDL clearance
- PMID: 18948618
- PMCID: PMC3426315
- DOI: 10.1161/CIRCRESAHA.108.183863
Mutation in EGFP domain of LDL receptor-related protein 6 impairs cellular LDL clearance
Abstract
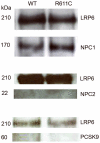
Mutation in the EGFP domain of LDL receptor-related protein 6 (LRP6(R611C)) is associated with hypercholesterolemia and early-onset atherosclerosis, but the mechanism by which it causes disease is not known. Cholesterol uptake was examined in cells from LRP6(+/-) mice and LRP6(R611C) mutation carriers. Splenic B cells of LRP6(+/-) mice have significantly lower LRP6 expression and low-density lipoprotein (LDL) uptake than those of the wild-type littermates. Although similar levels of total LRP6 were found in lymphoblastoid cells (LCLs) of LRP6(R611C) mutation carriers and those of the unaffected family member, LDL uptake was significantly lower in the mutant cells. Mutant and wild-type receptors show similar affinities for apolipoprotein B at neutral pH. LRP6 colocalized with LDL and was coimmunoprecipitated with NPC1 (Niemann-Pick disease type C1), an endocytic regulator of LDL trafficking. However, the cellular localization of LRP6 in the mutant cells shifted from cell surface to late endosomes/lysosomes. Plasma membrane expression levels of LRP6(R611C) was lower compared to wild-type receptor and declined to a greater extent in LDL-rich medium. Further examinations revealed lower efficacy of apolipoprotein B dissociation from LRP6(R611C) compared to wild-type receptor at an acidic pH. These studies identify LRP6 as a receptor for LDL endocytosis and imply that R611C mutation results in reduced LRP6 membrane expression and decreased LDL clearance. Based on our findings, we conclude that the increased affinity of the mutant receptor for LDL in acidic pH leads to their impaired dissociation in late endosomes, which compromises their recycling to the plasma membrane.
Figures















Similar articles
-
LRP6 protein regulates low density lipoprotein (LDL) receptor-mediated LDL uptake.J Biol Chem. 2012 Jan 6;287(2):1335-44. doi: 10.1074/jbc.M111.295287. Epub 2011 Nov 28. J Biol Chem. 2012. PMID: 22128165 Free PMC article.
-
ABCA1-dependent mobilization of lysosomal cholesterol requires functional Niemann-Pick C2 but not Niemann-Pick C1 protein.Biochim Biophys Acta. 2012 Mar;1821(3):396-404. doi: 10.1016/j.bbalip.2011.11.013. Epub 2011 Dec 10. Biochim Biophys Acta. 2012. PMID: 22179027
-
Wild-type LRP6 inhibits, whereas atherosclerosis-linked LRP6R611C increases PDGF-dependent vascular smooth muscle cell proliferation.Proc Natl Acad Sci U S A. 2011 Feb 1;108(5):1914-8. doi: 10.1073/pnas.1019443108. Epub 2011 Jan 18. Proc Natl Acad Sci U S A. 2011. PMID: 21245321 Free PMC article.
-
The combined hyperlipidemia caused by impaired Wnt-LRP6 signaling is reversed by Wnt3a rescue.Cell Metab. 2014 Feb 4;19(2):209-20. doi: 10.1016/j.cmet.2013.11.023. Cell Metab. 2014. PMID: 24506864 Free PMC article. Review.
-
Low-Density Lipoprotein Receptor-Related Protein 6 (LRP6) Is a Novel Nutritional Therapeutic Target for Hyperlipidemia, Non-Alcoholic Fatty Liver Disease, and Atherosclerosis.Nutrients. 2015 Jun 3;7(6):4453-64. doi: 10.3390/nu7064453. Nutrients. 2015. PMID: 26046396 Free PMC article. Review.
Cited by
-
Wnt Signaling in Atherosclerosis: Mechanisms to Therapeutic Implications.Biomedicines. 2024 Jan 25;12(2):276. doi: 10.3390/biomedicines12020276. Biomedicines. 2024. PMID: 38397878 Free PMC article. Review.
-
The role of Wnt signalling in development of coronary artery disease and its risk factors.Open Biol. 2020 Oct;10(10):200128. doi: 10.1098/rsob.200128. Epub 2020 Oct 21. Open Biol. 2020. PMID: 33081636 Free PMC article. Review.
-
Wnt-Lrp5 signaling regulates fatty acid metabolism in the osteoblast.Mol Cell Biol. 2015 Jun 1;35(11):1979-91. doi: 10.1128/MCB.01343-14. Epub 2015 Mar 23. Mol Cell Biol. 2015. PMID: 25802278 Free PMC article.
-
The LRP6 rs2302685 polymorphism is associated with increased risk of myocardial infarction.Lipids Health Dis. 2014 Jun 7;13:94. doi: 10.1186/1476-511X-13-94. Lipids Health Dis. 2014. PMID: 24906453 Free PMC article.
-
Mutant LRP6 Impairs Endothelial Cell Functions Associated with Familial Normolipidemic Coronary Artery Disease.Int J Mol Sci. 2016 Jul 22;17(7):1173. doi: 10.3390/ijms17071173. Int J Mol Sci. 2016. PMID: 27455246 Free PMC article.
References
-
- Llorente-Cortes V, Martinez-Gonzalez J, Badimon L. LDL receptor-related protein mediates uptake of aggregated LDL in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2000;20:1572–9. - PubMed
-
- Magoori K, Kang MJ, Ito MR, Kakuuchi H, Ioka RX, Kamataki A, Kim DH, Asaba H, Iwasaki S, Takei YA, Sasaki M, Usui S, Okazaki M, Takahashi S, Ono M, Nose M, Sakai J, Fujino T, Yamamoto TT. Severe hypercholesterolemia, impaired fat tolerance, and advanced atherosclerosis in mice lacking both low density lipoprotein receptor-related protein 5 and apolipoprotein E. J Biol Chem. 2003;278:11331–6. - PubMed
-
- Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, Kim DH, Ioka RX, Ono M, Tomoyori H, Okubo M, Murase T, Kamataki A, Yamamoto J, Magoori K, Takahashi S, Miyamoto Y, Oishi H, Nose M, Okazaki M, Usui S, Imaizumi K, Yanagisawa M, Sakai J, Yamamoto TT. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci U S A. 2003;100:229–34. - PMC - PubMed
-
- Beisiegel U, Weber W, Ihrke G, Herz J, Stanley KK. The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature. 1989;341:162–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

