PharmGKB: understanding the effects of individual genetic variants
- PMID: 18949600
- PMCID: PMC2677552
- DOI: 10.1080/03602530802413338
PharmGKB: understanding the effects of individual genetic variants
Abstract
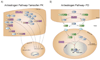
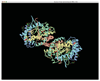
The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB: http://www.pharmgkb.org) is devoted to disseminating primary data and knowledge in pharmacogenetics and pharmacogenomics. We are annotating the genes that are most important for drug response and present this information in the form of Very Important Pharmacogene (VIP) summaries, pathway diagrams, and curated literature. The PharmGKB currently contains information on over 500 drugs, 500 diseases, and 700 genes with genotyped variants. New features focus on capturing the phenotypic consequences of individual genetic variants. These features link variant genotypes to phenotypes, increase the breadth of pharmacogenomics literature curated, and visualize single-nucleotide polymorphisms on a gene's three-dimensional protein structure.
Conflict of interest statement
Figures



References
-
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10(12):980. - PubMed
-
- Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res. Treat. 2007;101(1):113–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
