Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis
- PMID: 18952600
- PMCID: PMC2610494
- DOI: 10.1074/jbc.M805061200
Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis
Abstract
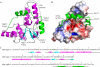
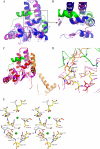
In prokaryotes, cognate toxin-antitoxin pairs have long been known, but no three-dimensional structure has been available for any given complex from Mycobacterium tuberculosis. Here we report the crystal structure and activity of a member of the VapBC family of complexes from M. tuberculosis. The toxin VapC-5 is a compact, 150 residues, two domain alpha/beta protein. Bent around the toxin is the VapB-5 antitoxin, a 33-residue alpha-helix. Assays suggest that the toxin is an Mg-enabled endoribonuclease, inhibited by the antitoxin. The lack of DNase activity is consistent with earlier suggestions that the complex represses its own operon. Furthermore, analysis of the interactions in the binding of the antitoxin to the toxin suggest that exquisite control is required to protect the bacteria cell from toxic VapC-5.
Figures


References
-
- Buts, L., Lah, J., Dao-Thi, M. H., Wyns, L., and Loris, R. (2005) Trends Biochem. Sci. 30 672-679 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

