A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis
- PMID: 18952777
- PMCID: PMC2590737
- DOI: 10.1105/tpc.108.061325
A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis
Abstract
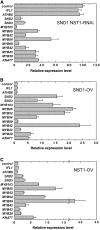
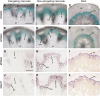
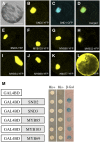
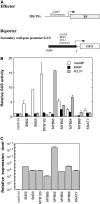
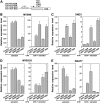
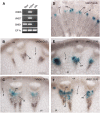
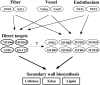
SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1) is a master transcriptional switch activating the developmental program of secondary wall biosynthesis. Here, we demonstrate that a battery of SND1-regulated transcription factors is required for normal secondary wall biosynthesis in Arabidopsis thaliana. The expression of 11 SND1-regulated transcription factors, namely, SND2, SND3, MYB103, MYB85, MYB52, MYB54, MYB69, MYB42, MYB43, MYB20, and KNAT7 (a Knotted1-like homeodomain protein), was developmentally associated with cells undergoing secondary wall thickening. Of these, dominant repression of SND2, SND3, MYB103, MYB85, MYB52, MYB54, and KNAT7 significantly reduced secondary wall thickening in fiber cells. Overexpression of SND2, SND3, and MYB103 increased secondary wall thickening in fibers, and overexpression of MYB85 led to ectopic deposition of lignin in epidermal and cortical cells in stems. Furthermore, SND2, SND3, MYB103, MYB85, MYB52, and MYB54 were able to induce secondary wall biosynthetic genes. Direct target analysis using the estrogen-inducible system revealed that MYB46, SND3, MYB103, and KNAT7 were direct targets of SND1 and also of its close homologs, NST1, NST2, and vessel-specific VND6 and VND7. Together, these results demonstrate that a transcriptional network consisting of SND1 and its downstream targets is involved in regulating secondary wall biosynthesis in fibers and that NST1, NST2, VND6, and VND7 are functional homologs of SND1 that regulate the same downstream targets in different cell types.
Figures







References
-
- Baudry, A., Heim, M.A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 366–380. - PubMed
-
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus, and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
-
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. - PubMed
-
- Burk, D.H., Zhong, R., Morrison, W.H.I.I.I., and Ye, Z.-H. (2006). Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J. Integr. Plant Biol. 48 85–98.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

